We are fascinated by the basic mechanisms of bacterial behavior and their evolution. We currently study a variety of processes central to bacterial growth, reproduction, cellular organization, adhesion, and biofilm formation. Our wide range of expertise and excellent collaborators allow us to focus on biological questions rather than specific methods and provides an outstanding multidisciplinary environment.
Our current areas of research activity include:
- Peptidoglycan synthesis in bacterial growth and division
- Mechanism and evolution of bacterial morphology
- Bacterial adhesion and biofilm formation
- Pili function, dynamics, an surface sensing
- Properties of bacterial adhesives
- Antimicrobial discovery
- Regulation of cell differentiation
- Control of cellular asymmetry and organization
- Evolutionary genomics
- Bacterial aging
See below for more details on these areas and links to a few key papers.
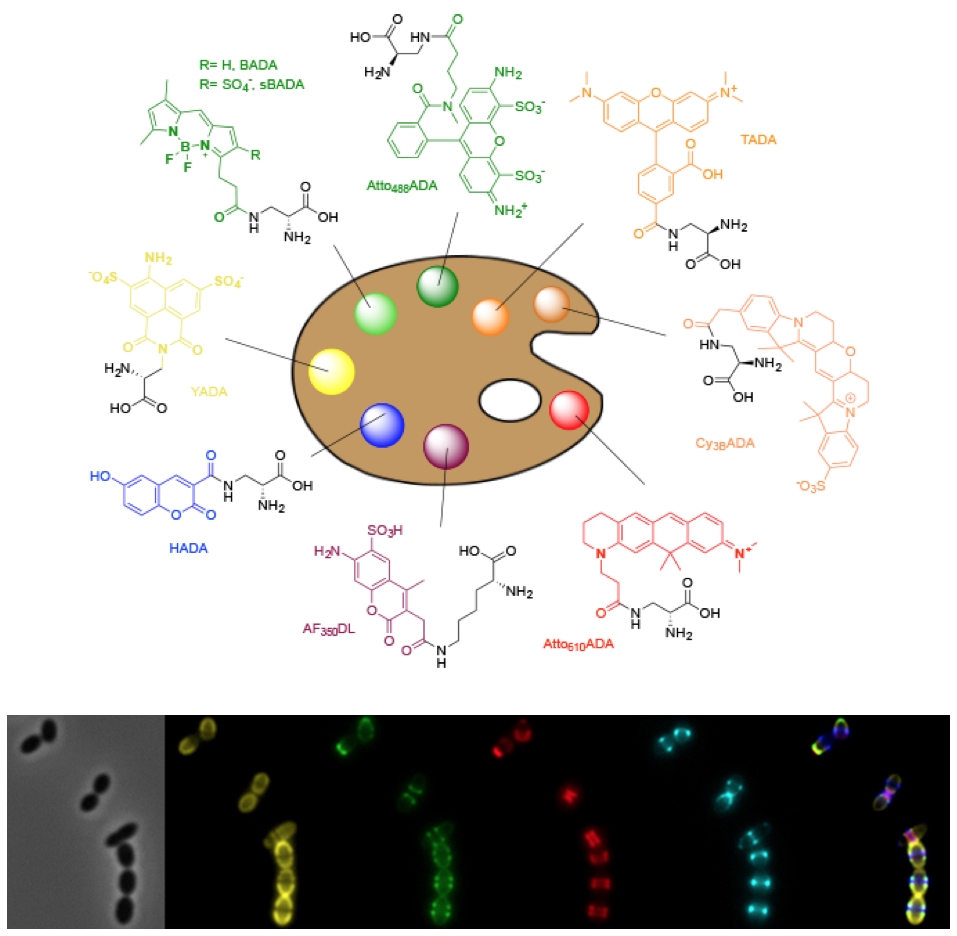
Peptidoglycan synthesis in bacterial growth and division
The spatial and temporal regulation of the synthesis of the peptidoglycan cell wall is critical for bacterial growth, division, and morphogenesis. We have developed fluorescent dyes, FDAAs, that label areas of peptidoglycan synthesis and have revolutionized its study. We use these tools to study the mechanisms of growth and division. Using an evolutionary biology approach, we have discovered that many bacterial species grow in unsuspected ways, and we are determining what variation in peptidoglycan synthesis mechanisms generate new modes of growth. See these articles: 1, 2, 3
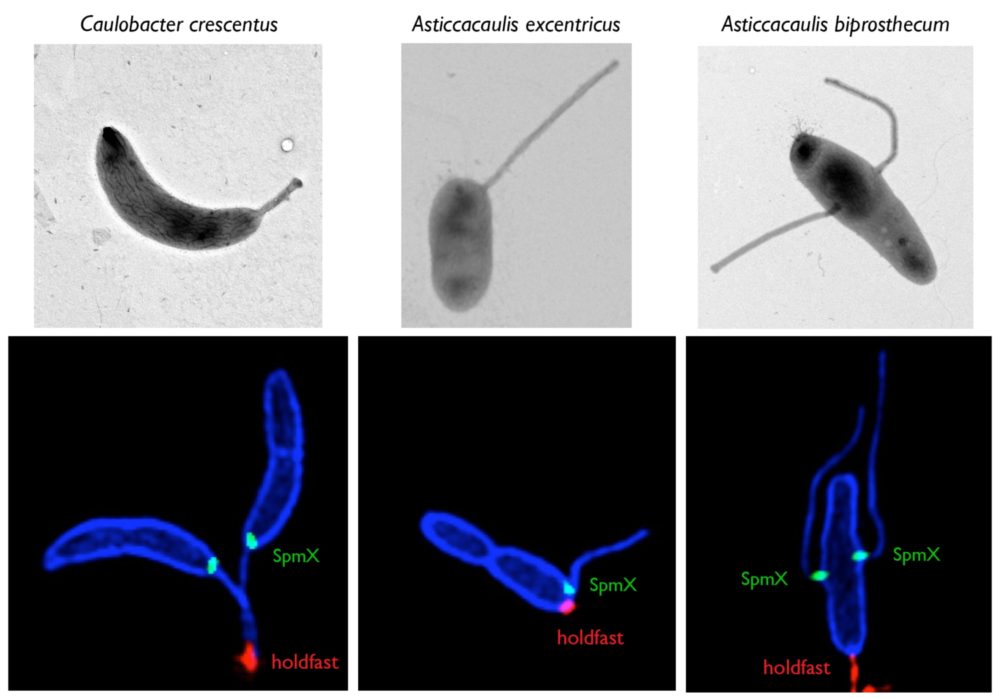
Mechanisms and evolution of bacterial morphology
Bacteria are found in a wide array of morphologies, yet understandings of bacterial shape are still almost exclusively sourced from research in rod-shaped model organisms. The modularity and plasticity of peptidoglycan synthesis and remodeling in different species remain mostly undefined. Our research of bacterial morphology has potential to uncover new aspects of bacterial fitness which are one of the most promising targets for antibiotic development. We use a combination of approaches, including state-of-the-art microscopy techniques, proteomics, comparative genomics, and forward genetic approaches to study the diversity of bacterial growth mechanisms and their evolution in Caulobacterales and Streptomyces species. See these articles: 1, 2, 3
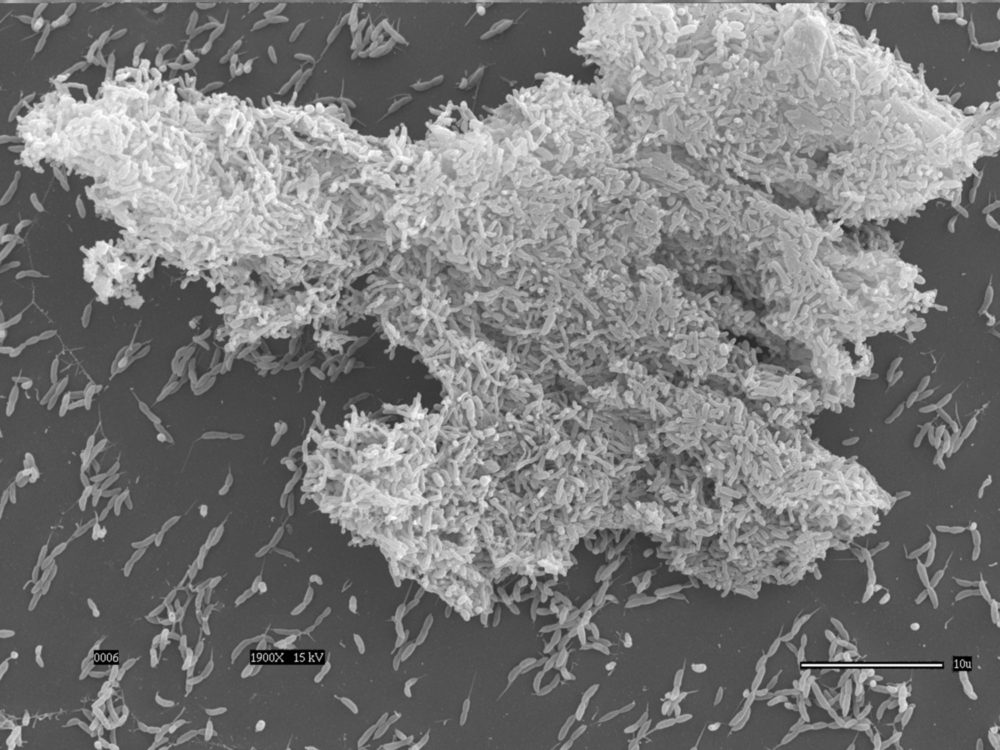
Bacterial adhesion and biofilm formation
Adhesion is an important component of bacterial infections and is the first step of biofilm formation. We study the mechanism and regulation of single cell attachment to surfaces, the synthesis of adhesins, and the subsequent events that lead to the formation of biofilms in Caulobacter crescentus. We also investigate the mechanisms by which cells sense surfaces to stimulate permanent adhesion, and the different factors that modulate production of holdfast, the adhesin Caulobacter produces to permanently adhere to surfaces. In addition, we study adhesion and biofilm formation in Hirshia baltica, a marine Caulobacterale, to understand how adhesins evolved to adapt to their environment. See these articles: 1, 2, 3
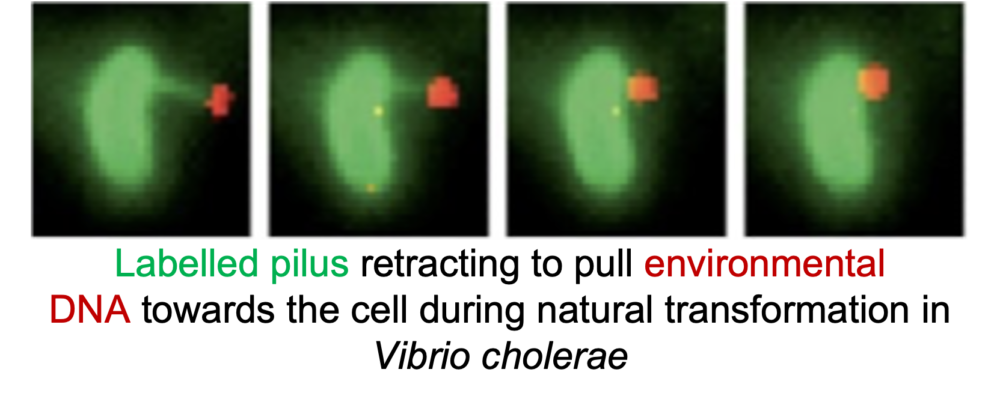
Pili function, dynamics, and surface sensing
Pili are ubiquitous bacterial nanomachines that facilitate many important cellular processes including surface adhesion, exchange of genetic material, and pathogenesis. We have developed labelling methods that permit the study of pilus dynamics in real time, as well as the means to physically block their retraction. Our current efforts are focused on deciphering the architecture of the type IVc, or tad, pilus nanomachine to identify and characterize the molecular determinants of pilus-mediated surface sensing and extension/retraction dynamics. See these articles: 1, 2, 3, 4

Properties of bacterial adhesives
Bioadhesives produced by bacteria are an abundant source of adhesives that offer impressive performance in their natural context, including on wet surfaces that remain a major problem for industrial adhesives. The so-called holdfast bioadhesive we study adheres to surfaces with one of the strongest adhesive forces in nature, matching the strongest industrial glues. We use a combination of genetic, biochemical, and biophysical methods to study the composition, structure, and properties of this impressive material. See these articles: 1, 2, 3
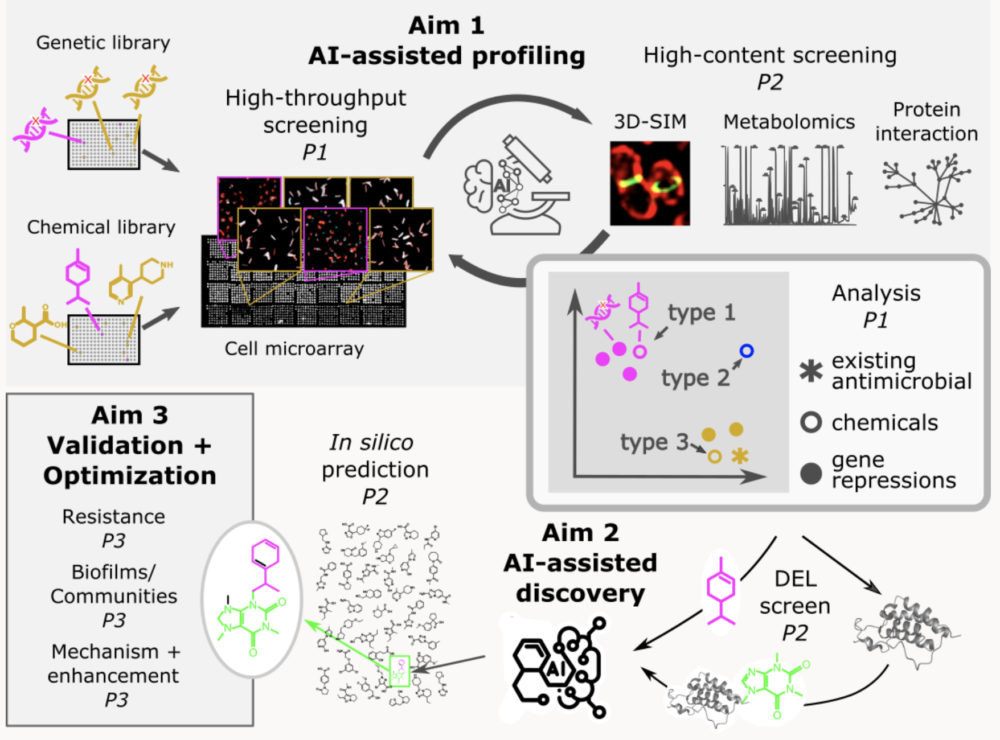
Antimicrobial discovery
We have collaborated with AI researchers to develop an AI-driven antimicrobial discovery strategy. An overview of our antimicrobial discovery strategy is presented in the figure. Gene repression and chemical libraries are screened comprehensively using high-throughput microscopy, with a subset of samples characterized in further detail using complementary high-content methods. The combined screening data forms the basis for developing an AI-based model of sample profiles for identifying various types of antimicrobials: those with novel mode of action either with (type 1) or without (type 2) an inferred target gene class; and those exploiting an existing mode of action (type 3) but possibly using an undiscovered mechanism. Based on these screening profiles and additional chemical affinity data from DNA-encoded chemical library (DEL) enrichment against desirable targets, an antimicrobial chemical prediction model will be adapted to search in silico, make-on-demand chemical databases. Compounds with high predicted antimicrobial activity will be obtained for detailed characterization and optimization.
Regulation of cell differentiation
The different stages of polar development in Caulobacter are tightly coordinated with the cell cycle. We study transcriptional and proteolytic regulatory mechanisms and regulators that control the timed and ordered progression of these stages. We recently discovered a novel mechanism of histidine kinase regulation through their dephosphorylation by a hub protein to control the timing of the cell cycle. See these articles: 1, 2, 3
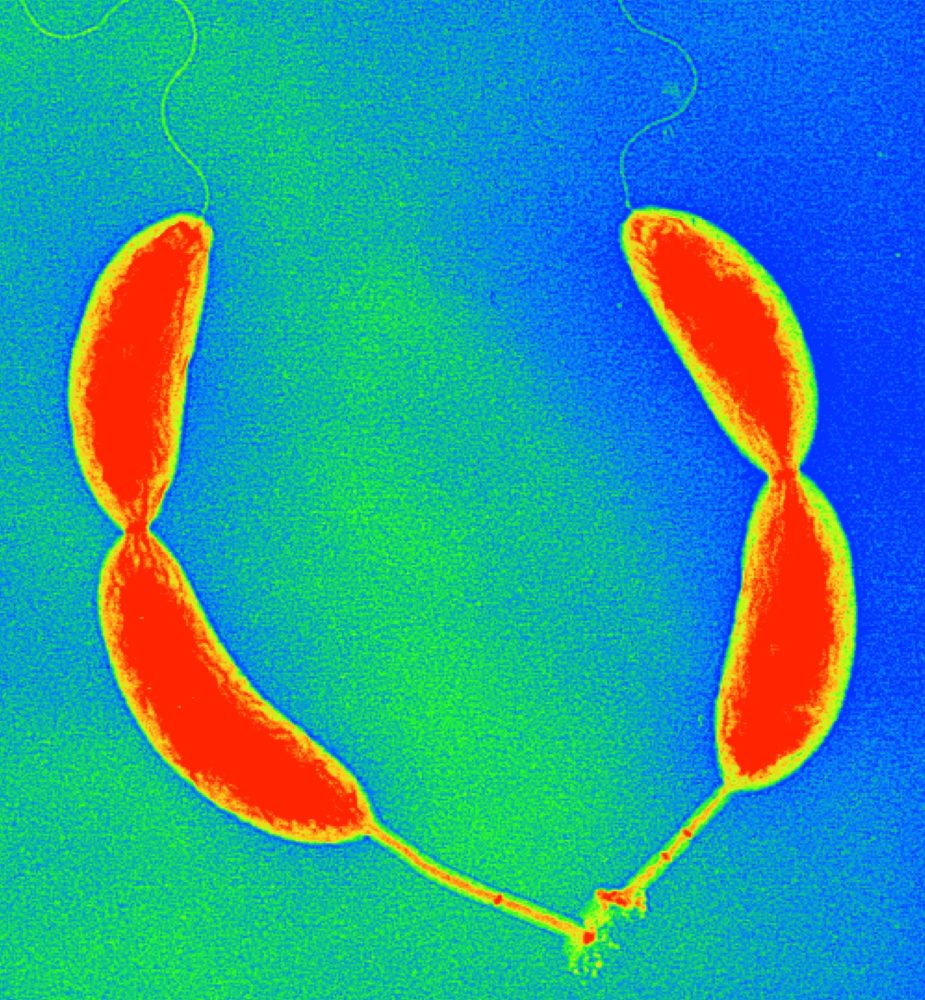
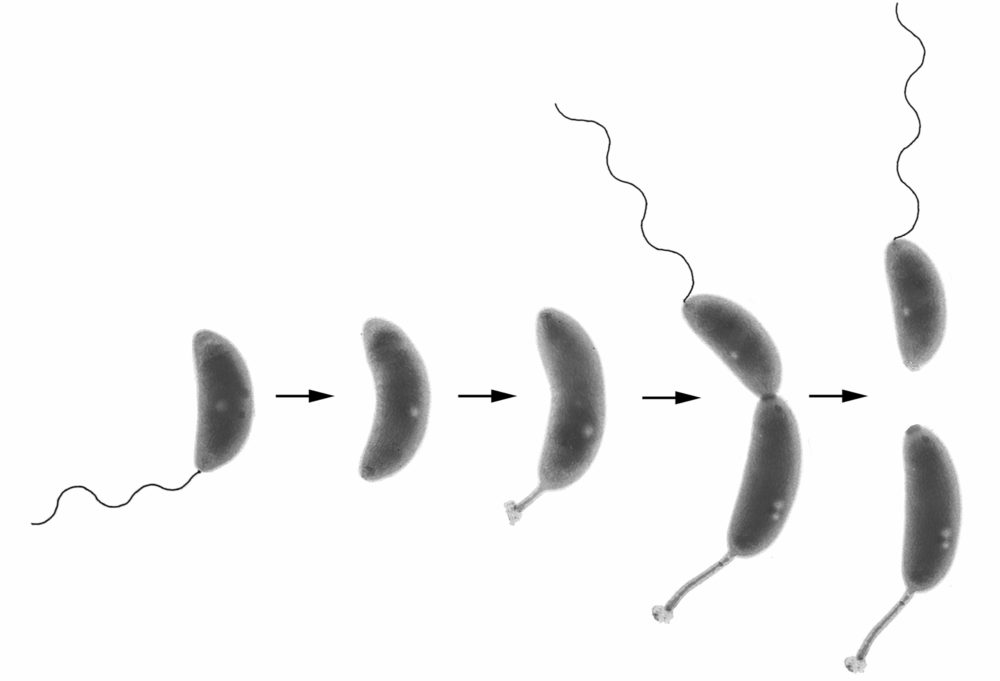
Control of cellular asymmetry and organization
Before they divide, Caulobacter cells are asymmetric with a flagellum at one pole and a stalk and adhesive holdfast at the other pole. We have used genetic and molecular methods to identify genes that are involved in the maintenance of cellular asymmetry and we are studying their mechanism of action. We also have a strong interest in the mechanisms that target proteins and structures to specific subcellular locations. See these articles: 1, 2, 3
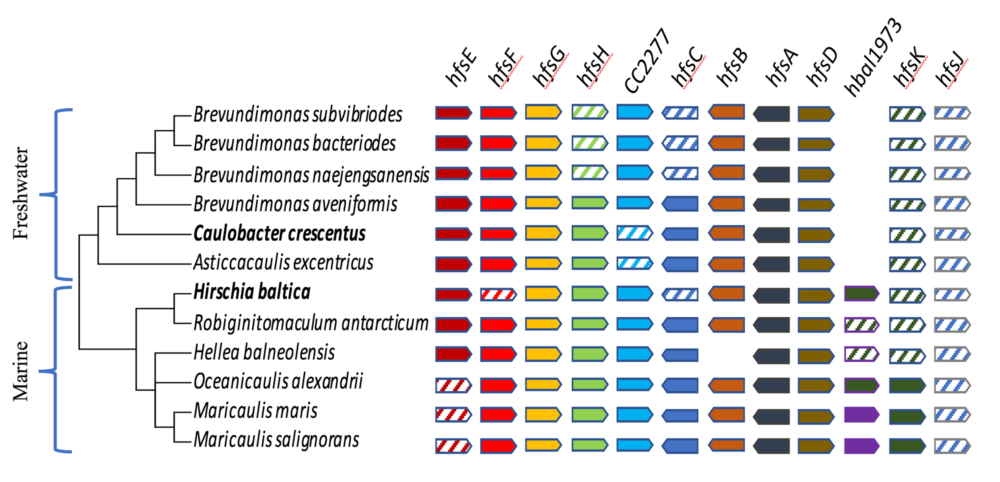
Evolutionary genomics
We take advantage of next generation sequencing methods to sequence the genome of bacteria closely related to Caulobacter to investigate the evolution of cell shape, cell differentiation, bacterial adhesion, and the regulatory networks that control them. Towards this goal, we are isolating bacteria related to Caulobacter from a variety of environments using a phylogeny based screen to avoid the bias due to isolations based on phenotype. See these articles: 1, 2, 3
Bacterial aging
Recent work has shown that organisms previously thought to be immortal, such as bacteria, demonstrate age-specific declines in reproduction. Asymmetry is thought to be important for aging by allowing the preferential segregation of damaged molecules to mother cells. We are using the asymmetrically dividing Caulobacter and close relatives that divide by the even more asymmetric mechanism of budding such as Hyphomonas to address the mechanism of aging and the role of asymmetry in this process. See these articles: 1, 2, 3
